Analysing a 6 TB planetary transcriptome with Spark
Recently, the Serratus dataset (Nature paper) was released by Robert C. Edgar et al. Serratus is a collection of transcriptomic (RNA) assemblies data-mined from NCBI SRA (Sequencing Read Archive), which has been used to discover novel virus genomes in existing data submissions. The authors describe the Serratus project as “accessing the planetary virome”. Very kindly, they provide all RNA assemblies from Serratus (not just viruses) to the public as a single file, hosted on Amazon S3, 6 TB uncompressed size. In the words of Rayan Chikhi, one of the researchers on the project:
This is possibly one of the largest, most diverse and easily accessible collections of transcriptome assemblies. The assemblies are draft quality, but they contain samples from soils, oceans, human microbiomes, entire mammalian transcriptomes, etc..
One way to investigate genomic datasets is k-mer analysis. K-mers are genomic fragments of a fixed length k, typically a small number < 60. They can serve as a building block for many other kinds of analysis. The most basic problem to solve with respect to k-mers is to count each distinct item, a problem known as k-mer counting.
We thought that the Serratus assemblies would be a useful technical challenge to improve the quality of our Discount k-mer counter, which runs on the Apache Spark framework. By solving a number of algorithmic and technical challenges - some relating to k-mer counting, some to Amazon Web Services (AWS), some to Spark, and some to Discount itself - we were able to k-mer count this entire dataset. For k = 28, it contains about 1.6 trillion distinct k-mers, and 6 trillion in total (full results at the end of this article). This also allowed us to validate our beliefs about the applicability of Spark to big genomic data.
Before we go into details, a disclaimer: JNP Solutions is not affiliated with the Serratus project and this article represents our own views and findings only, unless otherwise stated.
The place of big data technologies in genomics
The Serratus dataset is an example of an ongoing trend: genomic data, and the associated analysis needs, is constantly growing. Bioinformatics and data analysis is a major cost in genomic projects. As of September 2021, NCBI SRA contained around 25.6 petabase pairs and the growth continues. To get value from these growing datasets, we have to be able to run computations on them efficiently, whether for a single dataset or for a meta-analysis like Serratus.
For large data, many important jobs can’t run on a single machine, no matter how well equipped that machine is. CPU cores, RAM, and I/O speed will always be limiting factors, which imposes a size limit, making analyses impractical or impossible. To get around this, these jobs are now instead distributed (i.e. run on multiple machines simultaneously). On some level, this is not new. For a long time, scientists have been running jobs in traditional queues on clusters. But an alternative has emerged: big data frameworks such as Hadoop and Apache Spark.
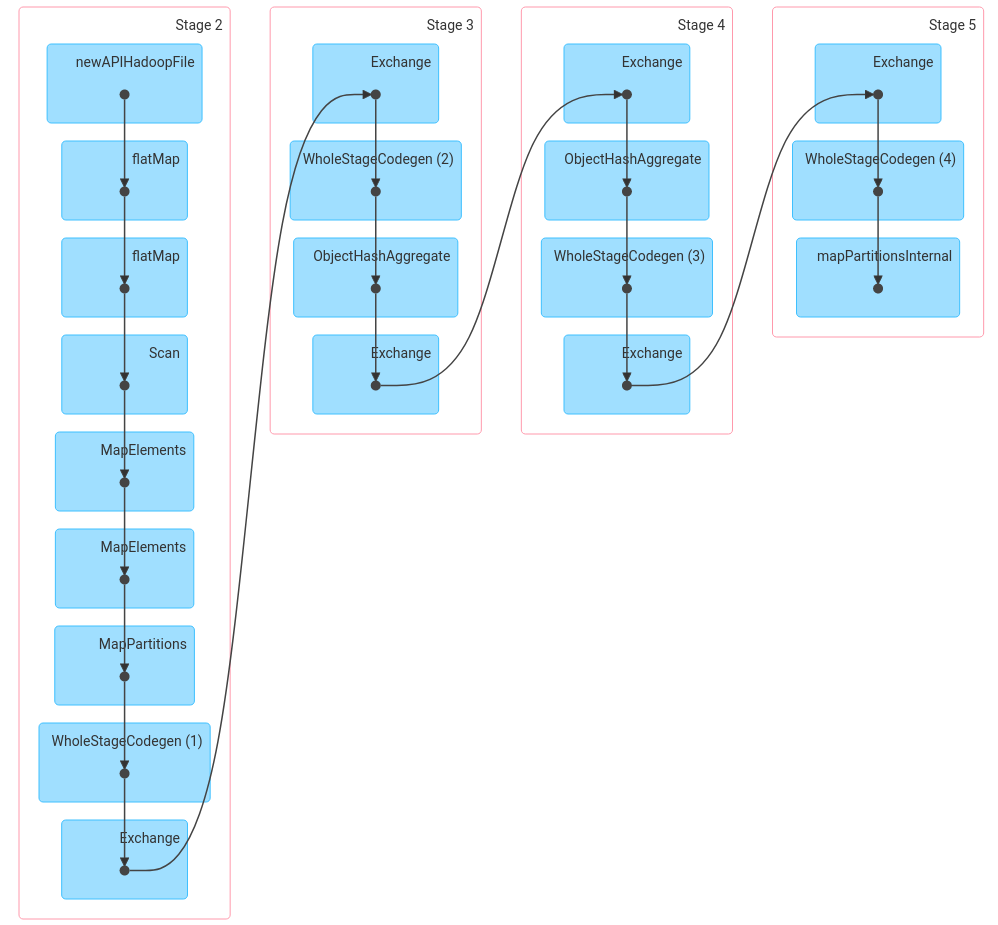
Spark brings several benefits to distributed computing, such as:
-
We can express the problem in a way that can be automatically optimised for a cluster, without having to worry about details of the individual machines. Multithreading is handled well, and data is kept in memory when possible, but “spilled” to disk if necessary.
-
Development cycles can be quick, thanks to support for interactive notebooks like Jupyter and Zeppelin and to the high-level API.
-
Spark scales to vast numbers of machines, even recovering automatically when machines fail or disappear. This is key to exploiting low-cost options such as spot pricing of cloud instances, where machines may sometimes be suddenly reassigned to other users when demand is high.
Finally, being able to easily take advantage of cloud infrastructure like AWS or GCP (Google Cloud Platform) can be a big benefit (although it is not required), since NCBI SRA is now being mirrored on these platforms. For these reasons, we take the view that in many cases, frameworks like Spark should be a preferred approach when developing new tools or pipelines for a distributed setting. They have the potential to unlock methods that we could not previously consider.
K-mer counting results
By applying Discount to this dataset, we were able to validate the usefulness of Spark with big genomic data in practice. After solving some technical problems, we were able to analyse the entire dataset. The statistical summary (k = 28, m = 13) is as follows.
==== Overall statistics ====
Number of buckets 10,399,311
Distinct k-mers 1,566,071,947,264
Unique k-mers 1,089,646,166,964
Total abundance 5,588,605,280,883
Mean abundance 3.569
Max abundance 139,750,690
Superkmer count 205,448,038,759
==== Per bucket (minimizer) statistics ====
Mean Min Max Std.dev
k-mers 150593.818 1 11111417 79471.415
abundance 537401.495 1 387870813 389447.442
superkmers 19755.928 1 6545965 14261.367
This makes the Serratus dataset the current scaling record for Discount, both in terms of the absolute data size and in terms of the number of distinct k-mers (complexity). To make this work smoothly, we had to do the following:
- Split the 6 TB uncompressed file into 60 parts of 100 GB each, to overcome a limitation of S3 and to be able to approach the dataset more gradually
- Improve support for large minimizers (m = 13) in Discount to obtain 10,000,000 buckets, decomposing complex data into small chunks
- Develop a new pre-grouped counting method that handles repetitive data efficiently through one extra Spark shuffle (a shuffle is a stage where all worker machines exchange data with each other)
- Tune partitioning and select AWS machine types to handle 18 TB shuffle data while maintaining close to 100% CPU utilisation
In the end, we were able to obtain a high degree of efficiency, which gave us the confidence that we would be able to scale to even larger data.
Conclusion
K-mer counting the massive Serratus dataset was an exciting challenge that forced us to develop several new features in Discount, as well as understand Spark and AWS better. The speed with which we were able to develop these new features and scale them up to large, previously unseen data also shows the value of Spark as a flexible, powerful and economical approach for big data jobs in genomics.
The new features developed for this project will be released as part of the upcoming Discount version 2.3. Our next steps with Serratus will be to go deeper into this dataset to gain biological insights, and to explore other applications of k-mer processing, such as metagenomic profiling.